The 2-Tissue (BFM) model implements fitting a two-tissue compartment model in each image pixel. It is based on an analytic solution of the system of differential equations which results in the calculation of two eigenvalues a1 and a2.
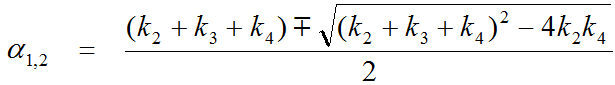
The expected tissue activity is obtained by the convolution of the input function with a sum of two decaying exponentials plus a contribution from whole blood.
![]()
For FDG, irreversible binding can be assumed and k4 set to zero, whereby a1 becomes zero. Hereby the number of fitted parameters is reduced and the operational equation simplifies to
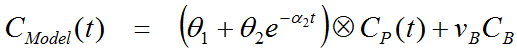
Overview of the BFM Processing in PXMOD
In the MRGlu (FDG BFM, Slice-dependent Times) model, sonly the irreversible configuration is supported. It uses the simplified basis function solution with fits 4 parameters: q1, q2, a2, vB. The operational equation is linear in the parameters q1, q2, vB, and nonlinear in a2 . The q1 and q2 parameters are a combination of the rate constants.
Basis function method according to Hong and Fryer [1] in the irreversible case:
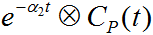 are pre-calculated for tabulated a2 values which span the prescribed range.
are pre-calculated for tabulated a2 values which span the prescribed range.It is possible to allow fitting of the blood volume fraction vB, or to fix it at a specific value. The configuration whether vB is fitted or fixed is specified in the preprocessing setup.
When setting up the processing it is recommended to enable calculation of the a2 map and inspect it regarding the prescribed range. If the prescribed maximum or minimum value is very frequently encountered this indicates that the range should be expanded.
Acquisition and Data Requirements
Image Data |
A dynamic FDG PET data set representing the measurements covering a sufficient time range after injecting of a 18F-Deoxy-Glucose (FDG) bolus. If the scanner FOV was not at a fixed location during the scan, the slice-times need to be contained in the image attributes. The model has been tested with the data of Siemens equipment (DICOM Conformance statement, see Frame Reference Time) capable of continuous table movement. |
Blood Data |
Blood activity from the time of injection until the end of the scan. It is important, that the blood and the image data have the same time base and decay correction is relative to the same time. |
Blood Preprocessing
The only necessary configuration is specification of the blood activity curve, either as a file, or as a VOI placed over a vessel such as the descending aorta or the left ventricle. Note, however, that the blood information must be available from the time of injection, whereas later measurements are sufficient for the tissue.
Model Preprocessing
The model processing panel includes two classes of options.
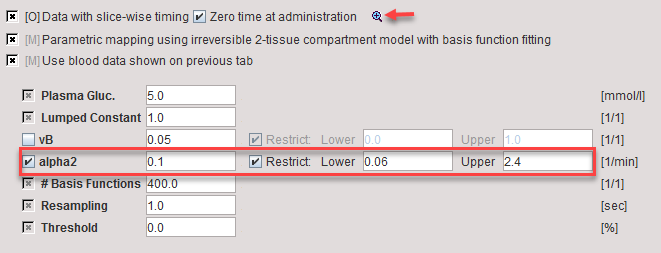
The check boxes at the top are related to the data:
Data with slice-wise timing |
Enabling the check instructs PXMOD to extract a dedicated time for each slice. |
Zero time at administration |
The slice reference times are relative to the time when the acquisition was started. If there is a delay between the tracer injection and scan start, this offset has to be added to the slice reference times. This operation is enabled by checking the box. Note, that if available an appropriated time is extracted from the Radiopharmaceutical Start DateTime (0018,1078) information. For GE data, the private field (0009,103B) is used. |
The indicated  button opens a dialog window for inspecting the times of the slices during the acquisitions.
button opens a dialog window for inspecting the times of the slices during the acquisitions.
The actual model parameters are listed in the lower part.
vB |
Blood volume fraction. Can be fitted or fixed. If checked, the blood fraction is fitted during map calculation. Otherwise, the specified value will be used for spillover correction. |
alpha2 |
Second eigenvalue. For defining its basis function range first enable the alpha2 parameter and then adjust the Lower and Upper values. |
#Basis |
Number of intermediate a2 values generated between Lower and Upper. The increments are logarithmically spaced. |
Resampling |
Sampling increment applied during the basis function calculation. |
Threshold |
Discrimination threshold for background masking. All pixels with energy below Threshold[%] of the maximal energy will be masked to zero. |
Model Configuration
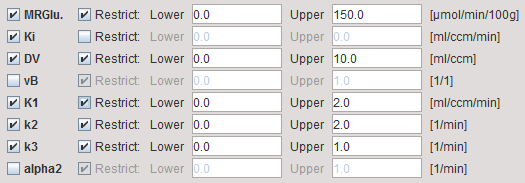
MRGlu |
Metabolic Rate of Glucose in [mmol/min/100ml], the actual result of the model. It is calculated as: |
Ki |
Influx Ki=(K1*k3)/(k2+k3) of the irreversible 2-tissue compartment model; is directly proportional to MRGlu. |
DV |
Distribution volume of FDG in tissue. |
vB |
Blood volume fraction defining the pixel-wise blood spillover correction. To fit, please activate in the Preprocessing tab. Note, however, that the noise will be increased. |
K1,k2,k3 |
Rate constants of the irreversible 2-tissue compartment model. |
alpha2 |
Shows the a2 values corresponding to the basis function of the solution. Can be used to check whether the defined range was adequate. |
Reference
1. Hong YT, Fryer TD: Kinetic modelling using basis functions derived from two-tissue compartmental models with a plasma input function: general principle and application to [18F]fluorodeoxyglucose positron emission tomography. Neuroimage 2010, 51(1):164-172. DOI