Texture analysis quantifies the variability of the pixel intensities within a structure such as a tumor. There is a plethora of metrics, which is discussed for instance by Hatt et al. [1] or Sollini et al. [2]including the formulae. The hope is that (some of) these metrics permit in vivo lesion characterization and provide predictive information about the tumor malignancy.
Rather than calculating hundreds of indexes, the texture facility implemented in PMOD provides a collection of 25 indexes which have been found to be most valuable in the context of PET imaging. Six are based on the pixel histogram within a VOI, and 19 based on the gray level co-occurrence matrix (GLCM).
Operation
Texture analysis is performed within the boundaries of a VOI which encloses the lesion of interest. Therefore, a first step for texture analysis consists of outlining the lesions to be analyzed by a suitable contouring technique. Once the VOIs are available and loaded on top of the images to be analyzed, texture analysis can be started by selecting the Texture analysis external tool as illustrated below.
A dialog window is opened which allows configuring the analysis parameters in the left part and shows information related to the texture calculation in the two panels to the right.
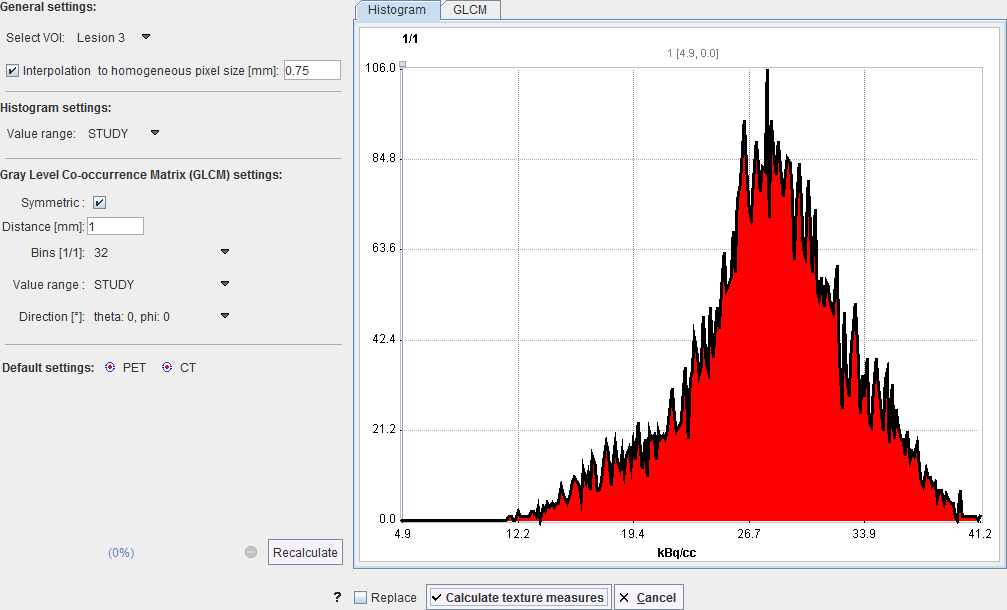
Select VOI allows choosing a representative VOI for which the information in the panels is updated. Interpolation to homogeneous pixel size is recommended for data which doesn't have isotropic resolution, and uses trilinear interpolation. Recalculate initiates calculation of the Histogram and the GLCM matrix for the VOI with the configured parameters and shows them for preview in the two panels. The actual calculation is started with the Calculate texture measures button
Histogram Analysis Configuration
The Histogram panel shows the histogram of the values within the selected VOI. The Value range selection defines whether the 256 bins cover the range of the STUDY, the current VOLUME, or just the VOI itself. This will be the input into the histogram-based analysis.
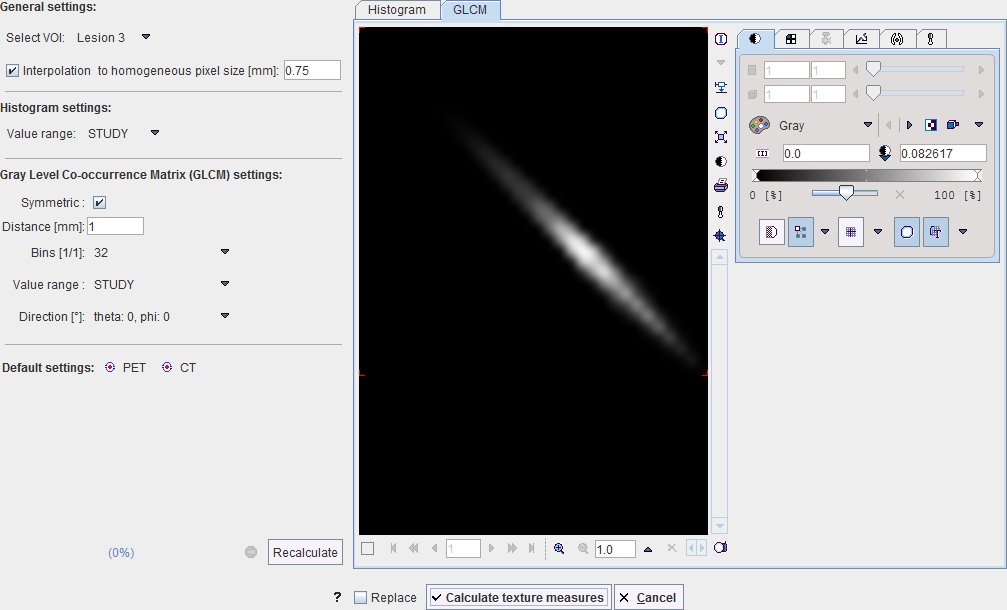
Gray Level Co-occurrence Matrix (GLCM) Analysis Configuration
The GLCM consists of the following steps to be configured on the Gray Level Co-occurrence Matrix settings:
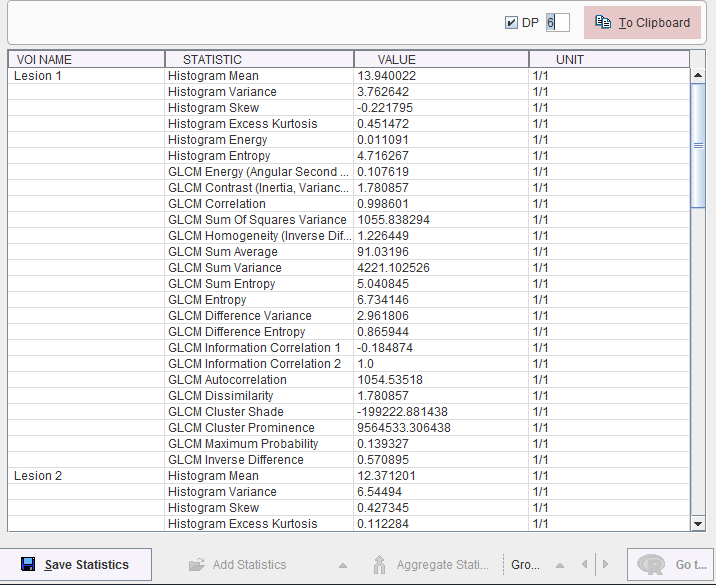
Texture Metrics
The metrics are calculated according to the formulae specified in the supplemental material of [1], please see there for explanations. They are obtained for all defined VOIs by activating the Calculate texture measures button, and are shown in a new dialog window, wehreas the configuration window is closed.
References
Hatt M, Tixier F, Pierce L, Kinahan PE, Le Rest CC, Visvikis D: Characterization of PET/CT images using texture analysis: the past, the present... any future? Eur J Nucl Med Mol Imaging 2017, 44(1):151-165. DOI
Sollini M, Cozzi L, Antunovic L, Chiti A, Kirienko M: PET Radiomics in NSCLC: state of the art and a proposal for harmonization of methodology. Scientific reports 2017, 7(1):358. DOI