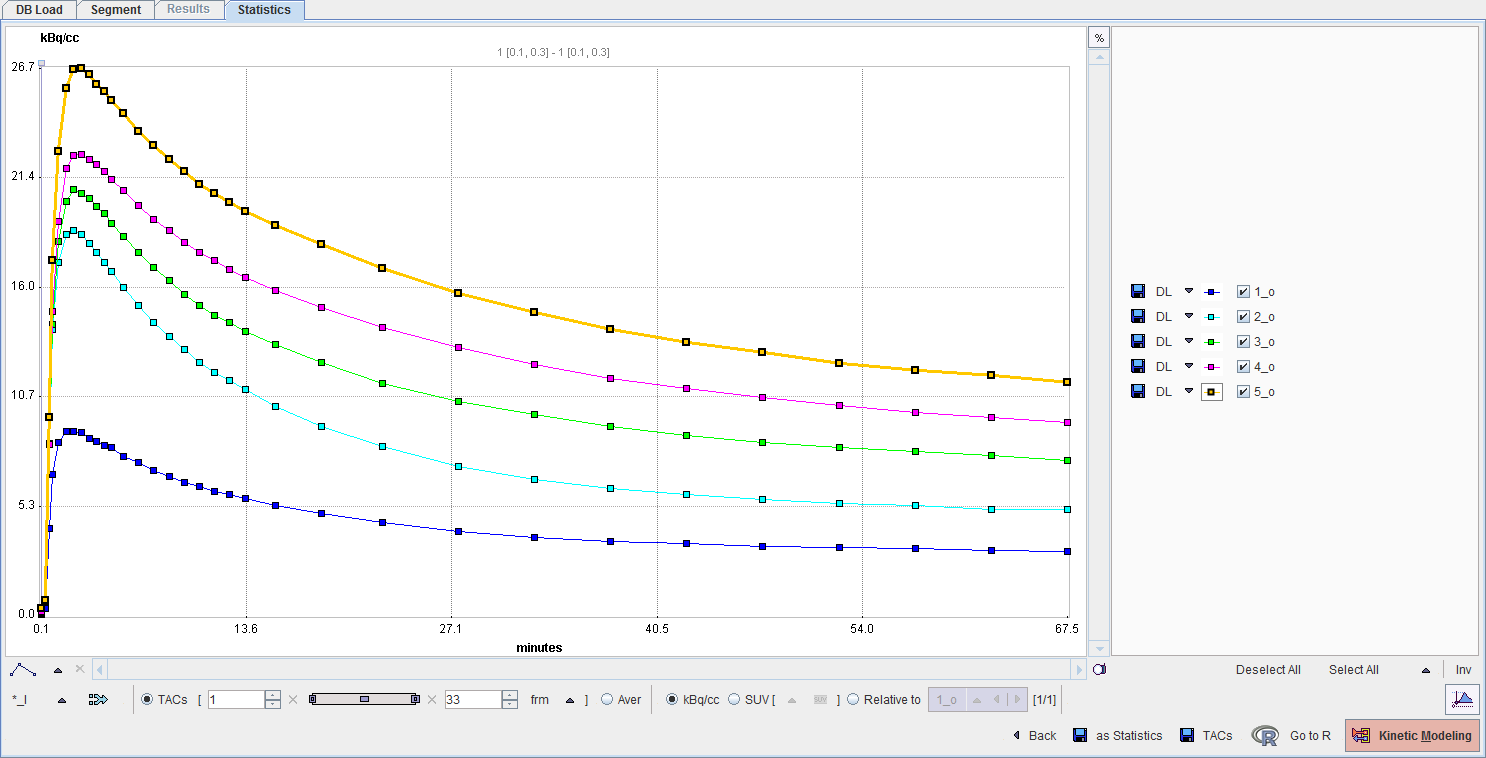
K-Means Cluster Analysis
The CLUSTER ANALYSIS method implements a simple k-means clustering procedure [1]. The Number of clusters (N) has to be defined, as well as the Number of iterations during which the clusters are adjusted. Note that all pixels within the body mask are assigned to one of the clusters, and the process doesn't include any kind of neighborhood relation. The classification is solely based on the signal time course in each pixel.
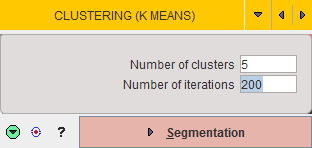
Segmentation
Once an appropriate body mask is ready and the segmentation parameters have been defined, please start calculations with the Segmentation button. The required processing time depends on several factors, particularly the number of iterations, and may take several minutes. The segmentation result is shown on the SEGMENTS page, initially in the layout shown below.
The display shows a fusion of the segmentation result (SEGMENTS) with the PET image as the reference.
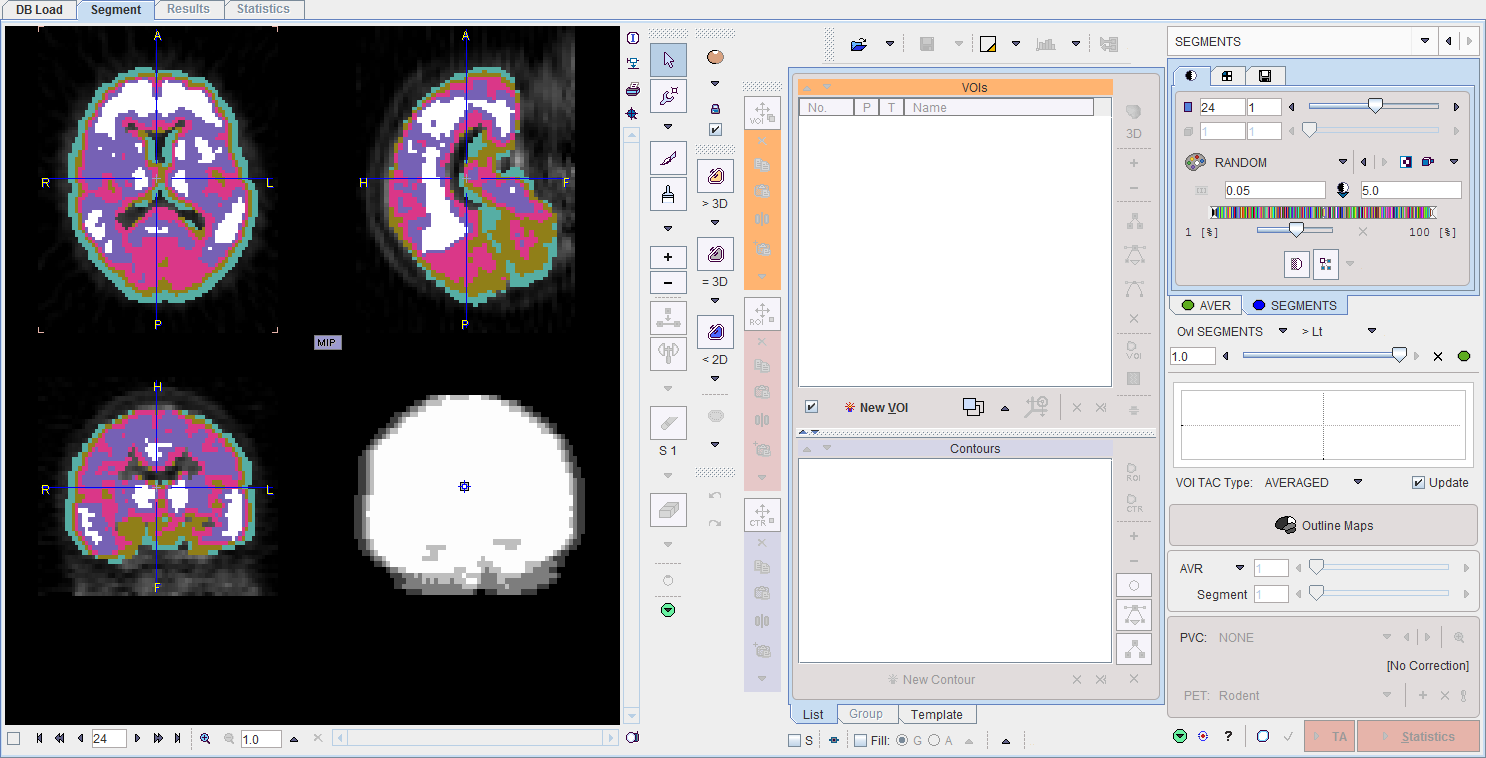
The procedures for converting clusters into VOIs are the same as for the ORGAN SEGMENTATION, except that the clustering only provides a single decomposition of the volume. Correspondingly, there is a static label images in the SEGMENTS tab rather than a dynamic series showing the volume in a hierarchical structure of decompositions, and the Segment slider is inactive.
All clusters can be outlined at once using the Outline Maps button, as illustrated below.
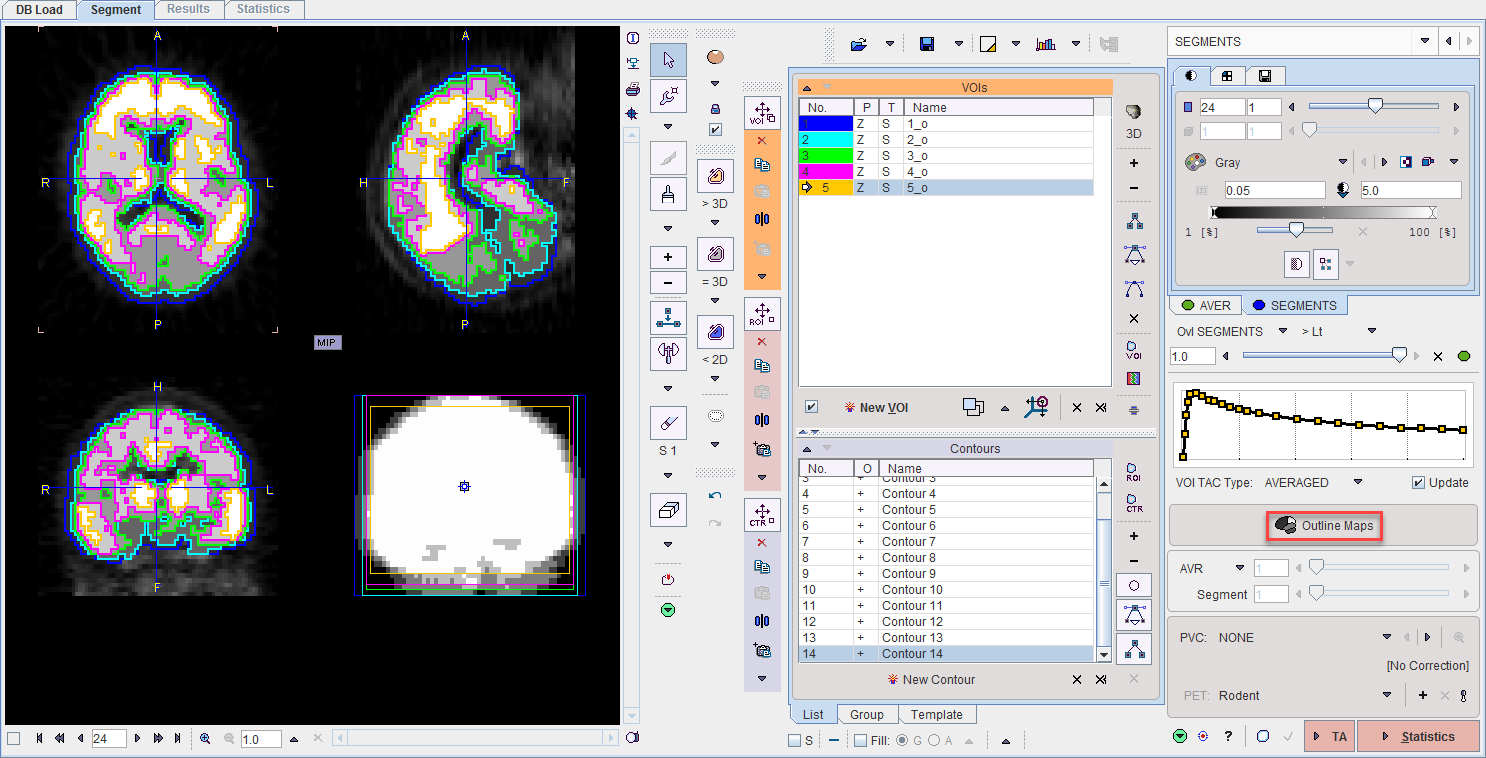
Finally, Statistics will calculate the average TACs of the clusters and show them on the Statistics page.